A GPU-Accelerated Fast Multipole Method for GROMACS: Performance and Accuracy | Journal of Chemical Theory and Computation

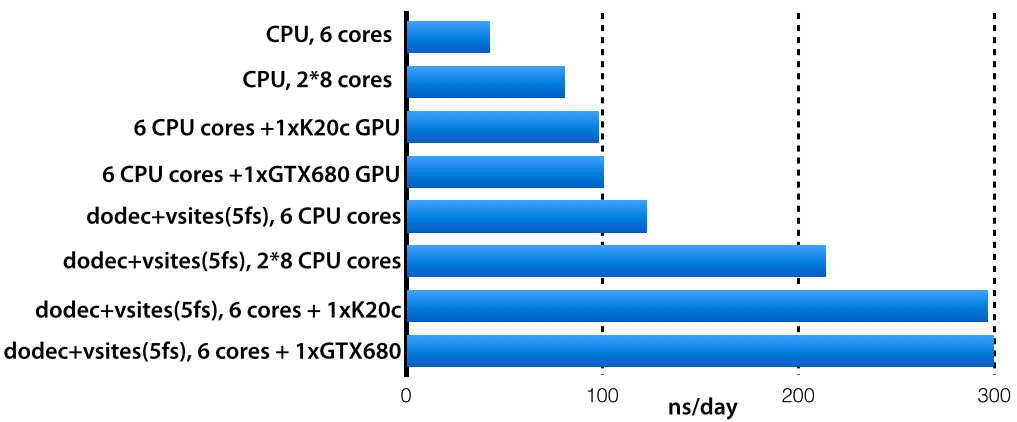
Comparison of job performance using GROMACS and ddCMD for one to four... | Download Scientific Diagram

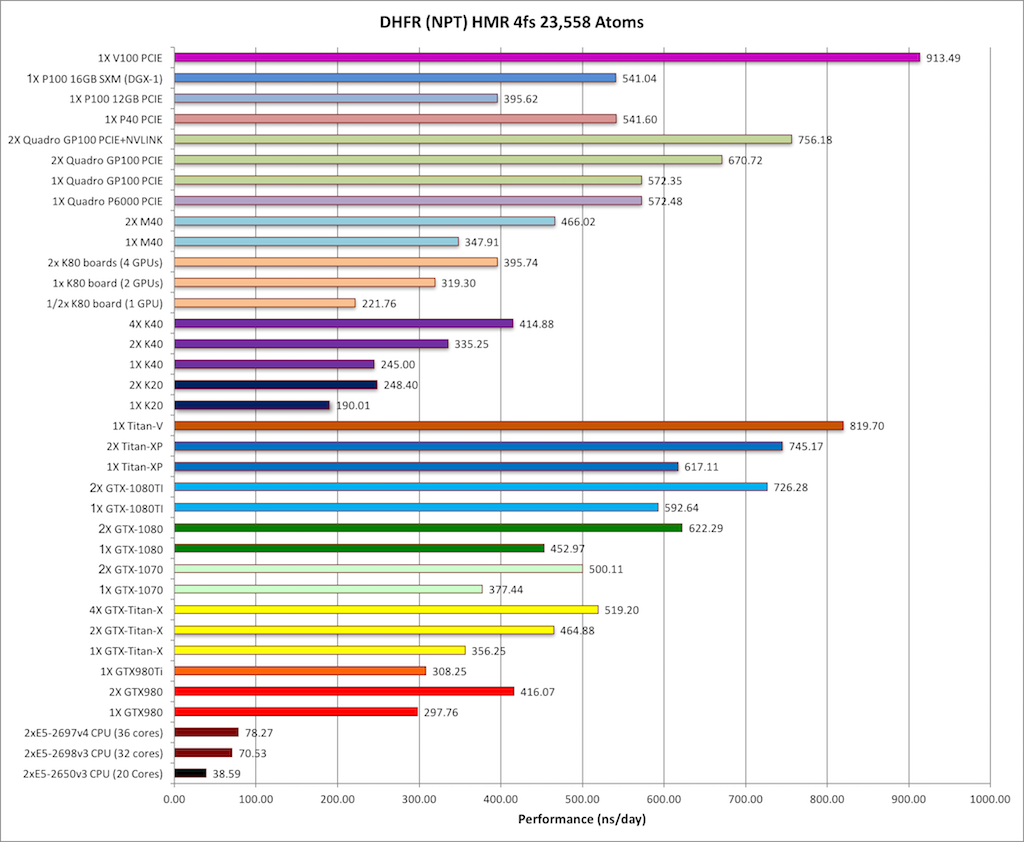
MDBenchmark: A toolkit to optimize the performance of molecular dynamics simulations: The Journal of Chemical Physics: Vol 153, No 14

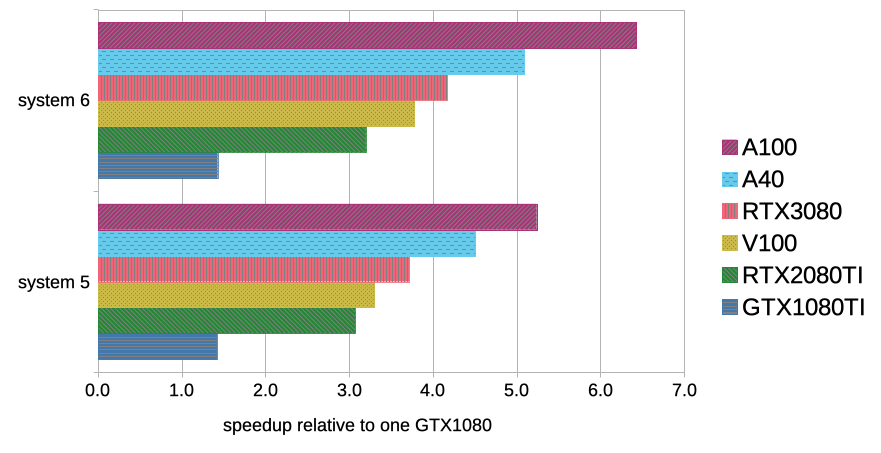
Proxy metric for P/P ratio of selected GPU models, computed as GROMACS... | Download Scientific Diagram

A GPU-Accelerated Fast Multipole Method for GROMACS: Performance and Accuracy | Journal of Chemical Theory and Computation

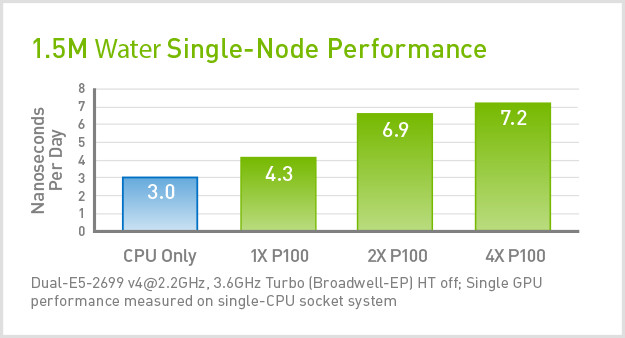
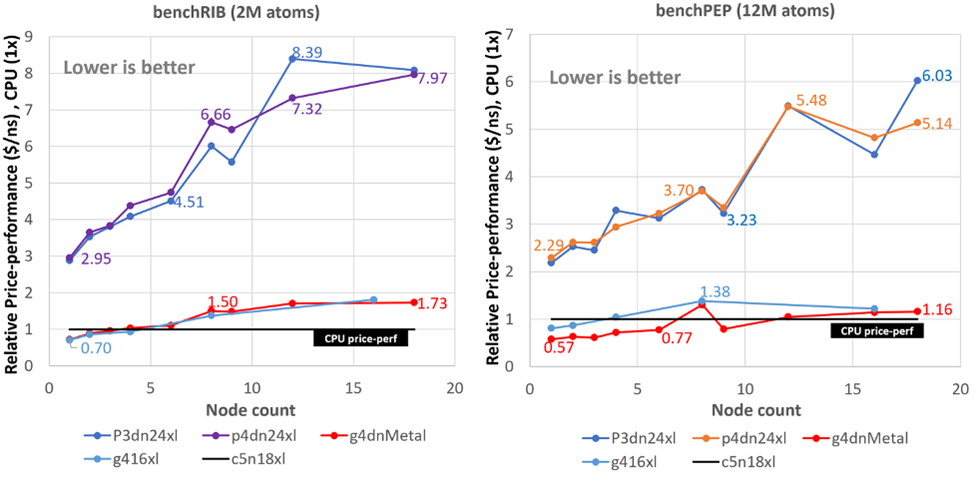
Webinar: More bang for your buck: Improved use of GPU Nodes for GROMACS 2018 (2019-09-05) – BioExcel – Centre of Excellence for Computation Biomolecular Research